Biography
Juan Luis Mateo Cerdán is Associate Professor in the Department of Computer Science at the University of Oviedo. His research is focused on Machine Learning and Data Mining with application to biological problems, specially in Genomics.
Before he was at the University of Heidelberg from 2010 to 2016. There, in the Centre for Organismal Studies, was part of Joachim Wittbrodt’s lab where he worked along with biologists applying his data analysis skills to different problems. At the same time he acquired an invaluable knowledge and experience in several kinds of molecular biology experiments.
His research and teaching activity began at the University of Castilla-La Mancha, in the Sistemas Inteligentes y Minería de Datos (SIMD) group led by José Antonio Gámez Marín and José Miguel Puerta Callejón.
- Bioinformatics
- Genomics
- Data Mining
- Machine Learning
-
PhD in Advanced Computer Technology, 2010
University of Castilla-La Mancha
-
Master in Advanced Computer Technology, 2008
University of Castilla-La Mancha
-
Engineering in Computer Science, 2002
University of Castilla-La Mancha
-
Technical Engineering in Computer Science, 2000
University of Castilla-La Mancha
Featured Publications
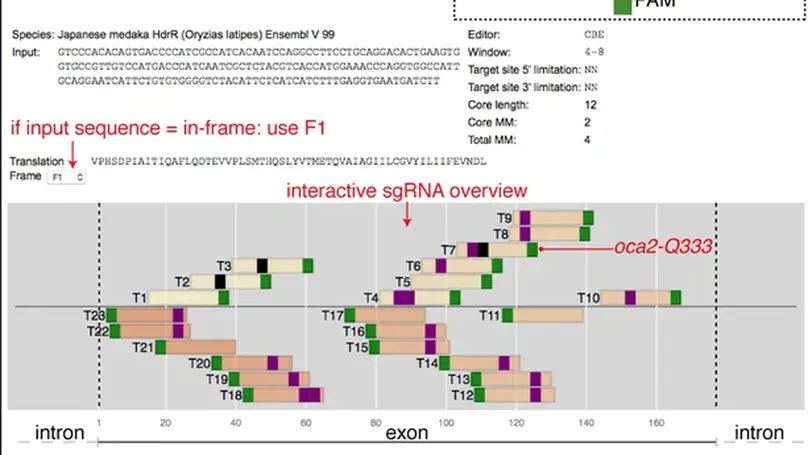
Single nucleotide variants (SNVs) are prevalent genetic factors shaping individual trait profiles and disease susceptibility. The recent development and optimizations of base editors, rubber and pencil genome editing tools now promise to enable direct functional assessment of SNVs in model organisms. However, the lack of bioinformatic tools aiding target prediction limits the application of base editing in vivo. Here, we provide a framework for adenine and cytosine base editing in medaka (Oryzias latipes) and zebrafish (Danio rerio), ideal for scalable validation studies. We developed an online base editing tool ACEofBASEs (a careful evaluation of base-edits), to facilitate decision-making by streamlining sgRNA design and performing off-target evaluation. We used state-of-the-art adenine (ABE) and cytosine base editors (CBE) in medaka and zebrafish to edit eye pigmentation genes and transgenic GFP function with high efficiencies. Base editing in the genes encoding troponin T and the potassium channel ERG faithfully recreated known cardiac phenotypes. Deep-sequencing of alleles revealed the abundance of intended edits in comparison to low levels of insertion or deletion (indel) events for ABE8e and evoBE4max. We finally validated missense mutations in novel candidate genes of congenital heart disease (CHD) dapk3, ube2b, usp44, and ptpn11 in F0 and F1 for a subset of these target genes with genotype-phenotype correlation. This base editing framework applies to a wide range of SNV-susceptible traits accessible in fish, facilitating straight-forward candidate validation and prioritization for detailed mechanistic downstream studies.
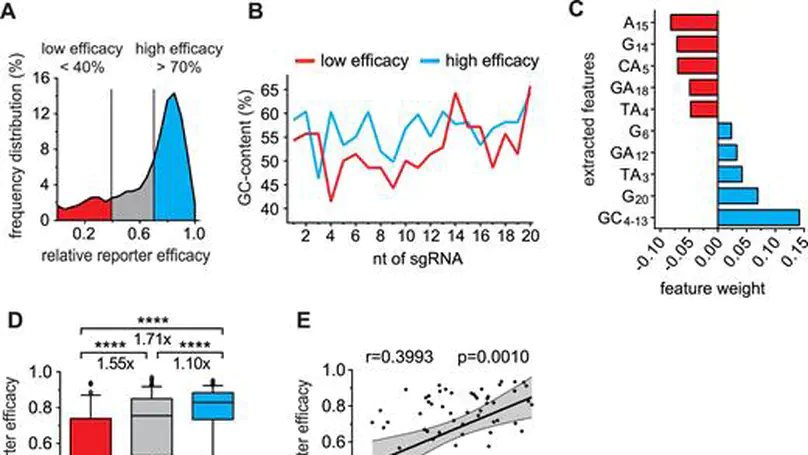
Genome editing with the CRISPR–Cas9 system has enabled unprecedented efficacy for reverse genetics and gene correction approaches. While off-target effects have been successfully tackled, the effort to eliminate variability in sgRNA efficacies—which affect experimental sensitivity—is in its infancy. To address this issue, studies have analyzed the molecular features of highly active sgRNAs, but independent cross-validation is lacking. Utilizing fluorescent reporter knock-out assays with verification at selected endogenous loci, we experimentally quantified the target efficacies of 430 sgRNAs. Based on this dataset we tested the predictive value of five recently-established prediction algorithms. Our analysis revealed a moderate correlation (r = 0.04 to r = 0.20) between the predicted and measured activity of the sgRNAs, and modest concordance between the different algorithms. We uncovered a strong PAM-distal GC-content-dependent activity, which enabled the exclusion of inactive sgRNAs. By deriving nine additional predictive features we generated a linear model-based discrete system for the efficient selection (r = 0.4) of effective sgRNAs (CRISPRater). We proved our algorithms’ efficacy on small and large external datasets, and provide a versatile combined on- and off-target sgRNA scanning platform. Altogether, our study highlights current issues and efforts in sgRNA efficacy prediction, and provides an easily-applicable discrete system for selecting efficient sgRNAs.
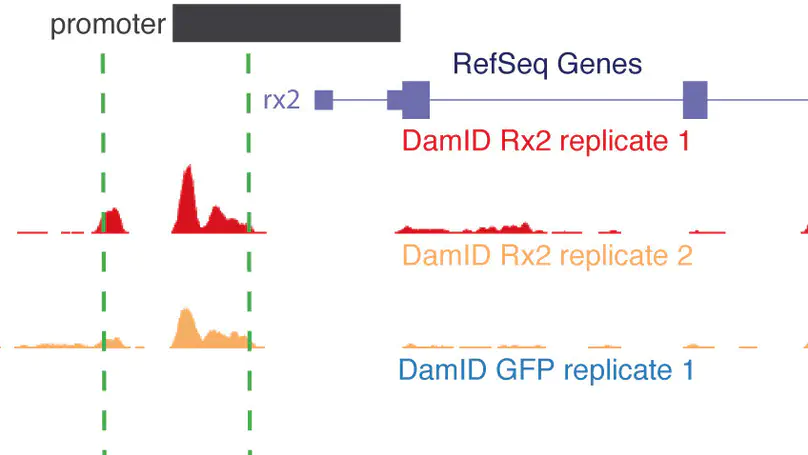
DNA adenine methyltransferase identification (DamID) has emerged as an alternative method to profile protein-DNA interactions; however, critical issues limit its widespread applicability. Here, we present iDamIDseq, a protocol that improves specificity and sensitivity by inverting the steps DpnI-DpnII and adding steps that involve a phosphatase and exonuclease. To determine genome-wide protein-DNA interactions efficiently, we present the analysis tool iDEAR (iDamIDseq Enrichment Analysis with R). The combination of DamID and iDEAR permits the establishment of consistent profiles for transcription factors, even in transient assays, as we exemplify using the small teleost medaka (Oryzias latipes). We report that the bacterial Dam-coding sequence induces aberrant splicing when it is used with different promoters to drive tissue-specific expression. Here, we present an optimization of the sequence to avoid this problem. This and our other improvements will allow researchers to use DamID effectively in any organism, in a general or targeted manner.
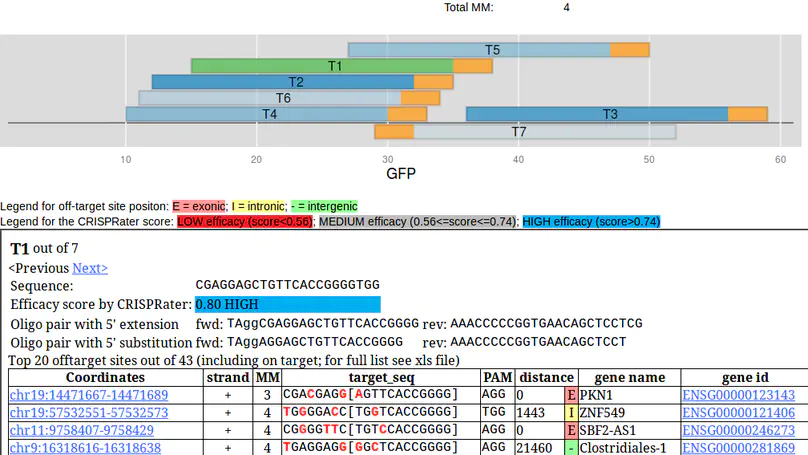
Engineering of the CRISPR/Cas9 system has opened a plethora of new opportunities for site-directed mutagenesis and targeted genome modification. Fundamental to this is a stretch of twenty nucleotides at the 5’ end of a guide RNA that provides specificity to the bound Cas9 endonuclease. Since a sequence of twenty nucleotides can occur multiple times in a given genome and some mismatches seem to be accepted by the CRISPR/Cas9 complex, an efficient and reliable in silico selection and evaluation of the targeting site is key prerequisite for the experimental success. Here we present the CRISPR/Cas9 target online predictor (CCTop, http://crispr.cos.uni-heidelberg.de) to overcome limitations of already available tools. CCTop provides an intuitive user interface with reasonable default parameters that can easily be tuned by the user. From a given query sequence, CCTop identifies and ranks all candidate sgRNA target sites according to their off-target quality and displays full documentation. CCTop was experimentally validated for gene inactivation, non-homologous end-joining as well as homology directed repair. Thus, CCTop provides the bench biologist with a tool for the rapid and efficient identification of high quality target sites.
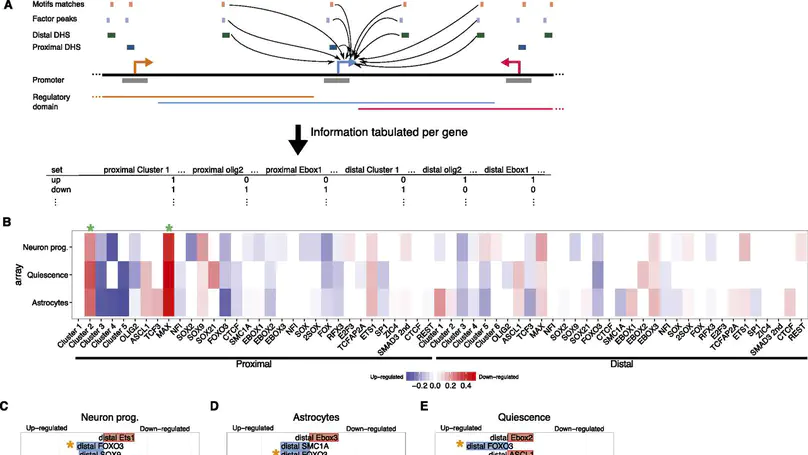
The gene regulatory network (GRN) that supports neural stem cell (NS cell) self-renewal has so far been poorly characterized. Knowledge of the central transcription factors (TFs), the noncoding gene regulatory regions that they bind to, and the genes whose expression they modulate will be crucial in unlocking the full therapeutic potential of these cells. Here, we use DNase-seq in combination with analysis of histone modifications to identify multiple classes of epigenetically and functionally distinct cis-regulatory elements (CREs). Through motif analysis and ChIP-seq, we identify several of the crucial TF regulators of NS cells. At the core of the network are TFs of the basic helix-loop-helix (bHLH), nuclear factor I (NFI), SOX, and FOX families, with CREs often densely bound by several of these different TFs. We use machine learning to highlight several crucial regulatory features of the network that underpin NS cell self-renewal and multipotency. We validate our predictions by functional analysis of the bHLH TF OLIG2. This TF makes an important contribution to NS cell self-renewal by concurrently activating pro-proliferation genes and preventing the untimely activation of genes promoting neuronal differentiation and stem cell quiescence.
Recent Publications
Software
ACEofBASEs (a careful evaluation of BaseEdits) standalone

ACEofBASEs is a tool to determine sites to be editted with the CRISPR/Cas9 technology in a input sequence and predict its potential off-target sites. The online version of ACEofBASEs is available at http://aceofbases.cos.uni-heidelberg.de/
The standalone version has a command line interface designed to allow search of large volume of sequences and higher flexibility. Also advisable if the target genome is not publicly available.
Access ACEofBASEs standalone from my bitbucket site.
CCTop (CRISPR/Cas9 target online predictor) standalone

CCTop is a tool to determine suitable CRISPR target sites in a given query sequence(s) and predict its potential off-target sites. The online version of CCTop is available at http://cctop.cos.uni-heidelberg.de/
The standalone version has a command line interface designed to allow search of large volume of sequences and higher flexibility. Also advisable if the target genome is not publicly available.
Access CCTop standalone from my bitbucket site.
iDEAR (iDamID Enrichment Analysis with R)
iDEAR identifies the binding profile of a transcription factor (or any DNA binding protein) from iDamIDseq data according to our publication in Development.
Access iDEAR from my bitbucket site.
Teaching
Currently I teach at the University of Oviedo the following subjects:
- Sistemas Inteligentes (Grado en Ingeniería Informática del Software)
- Programming Methodology (Grado en Ingeniería Informática en Tecnologías de la Información)
- Bioinformática (Máster Universitario en Biotecnología Aplicada a la Conservación y Gestión Sostenible de Recursos Vegetales)
Previously
- Repositorios de Información (Grado en Ingeniería Informática del Software)
- Sistemas Operativos (Grado en Ingeniería Informática en Tecnologías de la Información)
- Tecnologías y Paradigmas de la Programación (Grado en Ingeniería Informática en Tecnologías de la Información)
- Arquitectura del Software (Grado en Ingeniería Informática del Software)
At the University of Heidelberg I’ve taught these subjects (2014-2016):
- Introduction to Statistical Analysis for Biologists with R (Bachelor in the Faculty of Biosciences)
- Data Analysis with R (Bachelor in the Faculty of Biosciences)
- Introduction to Genetics (Bachelor in the Faculty of Biosciences)
- Statistics for Biology: design, analysis and visualization (Core course in the HBIGS International PhD Program in Molecular and Cellular Biology)
At the University of Castilla-La Mancha I taught these subjects (2006-2010):
- Minería de Datos (Ingeniería Informática)
- Bioinformática (Ingeniería Informática)
- Metodología de la Programación (Ingeniería Informática)
- Estructuras de Datos y de la Información (Ingeniería Informática)
Contact
- +34 985 10 2943
- Jesús Arias de Velasco s/n, 33005 Oviedo, Spain
- Geology Faculty, 7th floor, Room 7-1