iDamIDseq and iDEAR: an improved method and computational pipeline to profile chromatin-binding proteins
Abstract
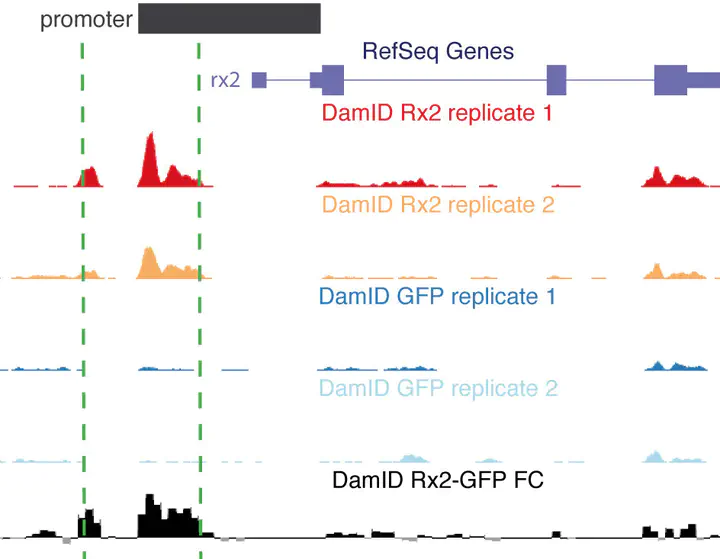
DNA adenine methyltransferase identification (DamID) has emerged as an alternative method to profile protein-DNA interactions; however, critical issues limit its widespread applicability. Here, we present iDamIDseq, a protocol that improves specificity and sensitivity by inverting the steps DpnI-DpnII and adding steps that involve a phosphatase and exonuclease. To determine genome-wide protein-DNA interactions efficiently, we present the analysis tool iDEAR (iDamIDseq Enrichment Analysis with R). The combination of DamID and iDEAR permits the establishment of consistent profiles for transcription factors, even in transient assays, as we exemplify using the small teleost medaka (Oryzias latipes). We report that the bacterial Dam-coding sequence induces aberrant splicing when it is used with different promoters to drive tissue-specific expression. Here, we present an optimization of the sequence to avoid this problem. This and our other improvements will allow researchers to use DamID effectively in any organism, in a general or targeted manner.